At this Monday’s Montreal R User Group meeting, Arthur Charpentier gave an interesting talk on the subject of quantile regression.
One of the main messages I took away from the workshop was that quantile regression can be used to determine if extreme events are becoming more extreme. The example given was hurricane intensity since 1978. It may be that the average intensity is not increasing and therefore a standard linear regression would show no trend (since linear regression predicts expected, or mean values), but that’s not really what we are interested in anyway. If we are going to formulate proper risk models, what we want to know is whether the strong hurricanes are getting stronger. This is where quantile regression comes in.
I always find that the best way for me to check my understanding is to simulate some data and check to see that things are behaving the way I expect them to. The advantage of doing this rather than just playing with data is that you know what the real process is, since you defined it. To get a handle on this stuff, I simulated some non-gausian (gamma distributed) data to mimic the hurricane data. I set it up so that the mean intensity stays constant across years, and the variance increasing constantly over time such that the intense (simulated) hurricanes get more intense over time.
## Simulate some non-gausian data with constant mean
## and increasing variance
n_i<-80
d<-array(dim=c(n_i*20,2))
for(i in 1:20)
{
d[((i-1)*n_i+1):(i*n_i),2]<-rgamma(n_i,i,i)
d[((i-1)*n_i+1):(i*n_i),1]<-21-i
}
plot(d)
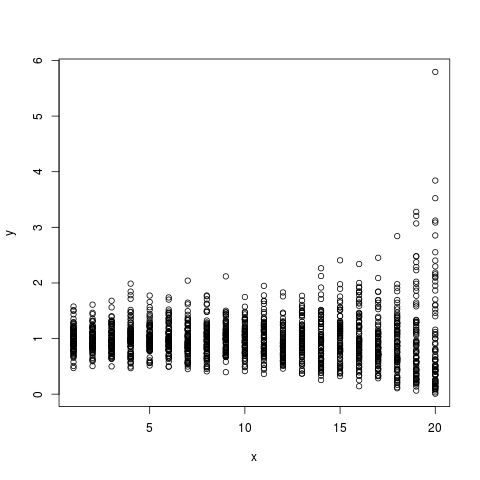
I then followed the procedure suggested by Arthur, which is to conduct quantile regressions across the quantile range (0,1). The results can be plotted as quantile vs the regression coefficient in order to see the magnitude and direction of the relationship across the quantile range.
## Run quantile regression on the simulated data
## across a range of quantiles
u=seq(.025,.975,by=.01)
coefstd=function(u) summary(rq(d[,2]~d[,1],tau=u))$coefficients[,2]
coefest=function(u) summary(rq(d[,2]~d[,1],tau=u))$coefficients[,1]
CS=Vectorize(coefstd)(u)
CE=Vectorize(coefest)(u)
## Plot the results
k=2
plot(u,CE[k,],type='l',xlab=expression(tau),ylab='Coefficient')
polygon(c(u,rev(u)),c(CE[k,]+1.96*CS[k,],rev(CE[k,]-1.96*CS[k,])),col='grey')
lines(u,CE[k,])
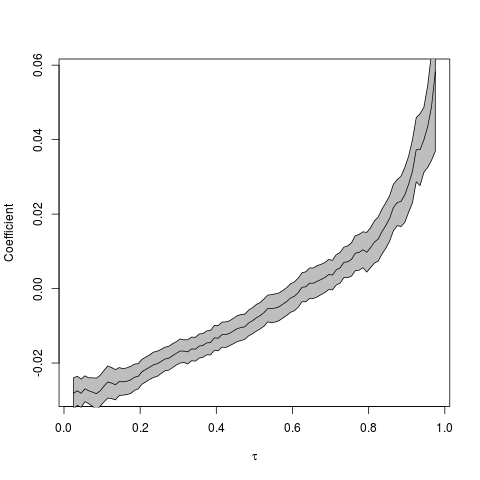
So, the coefficient seems to be an increasing function of the quantile (tau). But how do we interpret this? Low intensity (simulated) storms are becoming less intense given that the regression coefficient at low quantiles is negative. More importantly, however, is that the high intensity (simulated) storms are becoming more intense. We can see this by noting that the regression coefficients in the high quantile range are positive, and increasing.
Another way to visualise the quantile regression results is by animating the regressions together to see how the relationship changes across the quantiles (tau).
Click the image to see the animated GIF.

Arthur has a prettier animation of this type using the actual hurricane data here.